Portfolio
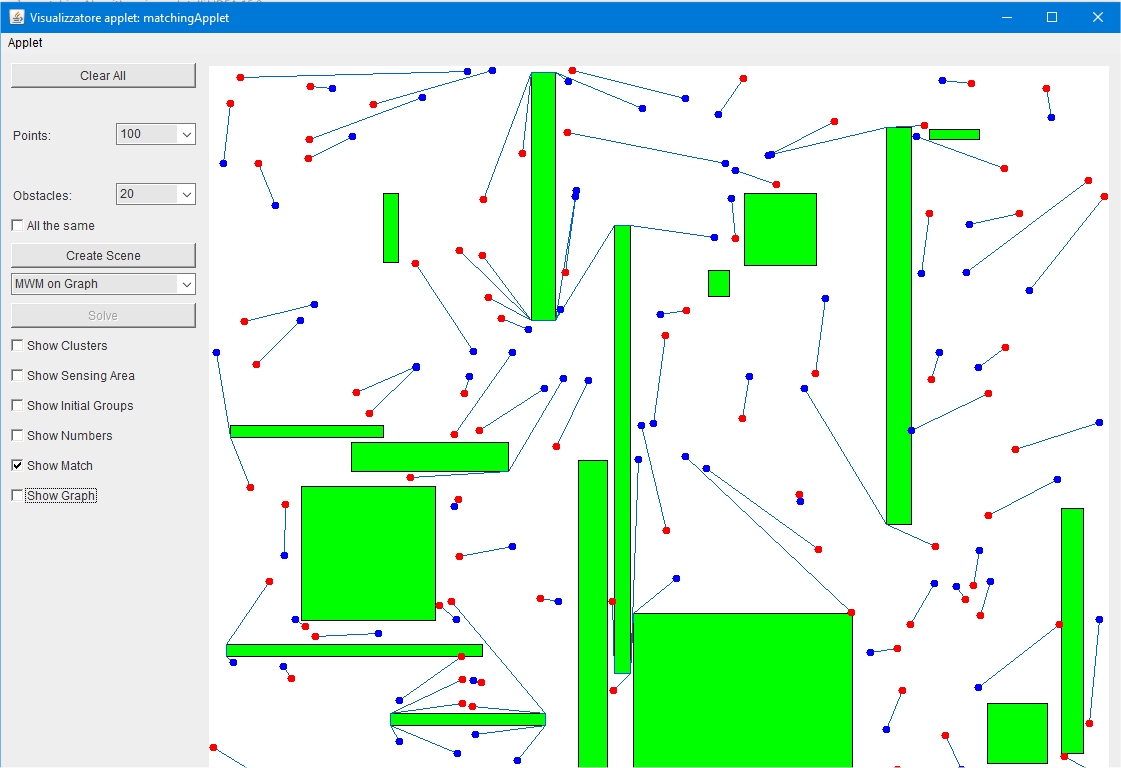
Matching Algorithms is a visual tool for computing visibility graphs and apply bipartite matching algoritms for a set of mobile stations that needs to move from a start position to a destination. In a real situation stations move in a relatively open space where obstacles are present. Obstacles may obstruct the path of the base stations when moving from one position to the next.
This tool has been developed with other Authors as a research project at University of Catania.
Matching Algoritms is supported by StudioPigola on all editions of Windows 10.
REFERENCES
- 2011 S. Cristaldi, A. Ferro, R. Giugno, G. Pigola, A. Pulvirenti, “Obstacles Constrained Group Mobility Models in Event-Driven Wireless Networks with Movable Base Stations”, Ad Hoc Networks 9 (2011), pp. 400-417, DOI information: 10.1016/j.adhoc.2010.07.004
- 2006 A. Ferro, R. Giugno, M. Mongiovi’, G. Pigola, A. Pulvirenti, “Distributed Clustering and Closest-Match Motion Planning Algorithms for Wireless Ad-Hoc Networks with Movable Base Stations”, Med-Hoc-Net 2006, (pp. 53-59). Isbn/Issn: 88-902405-1-2
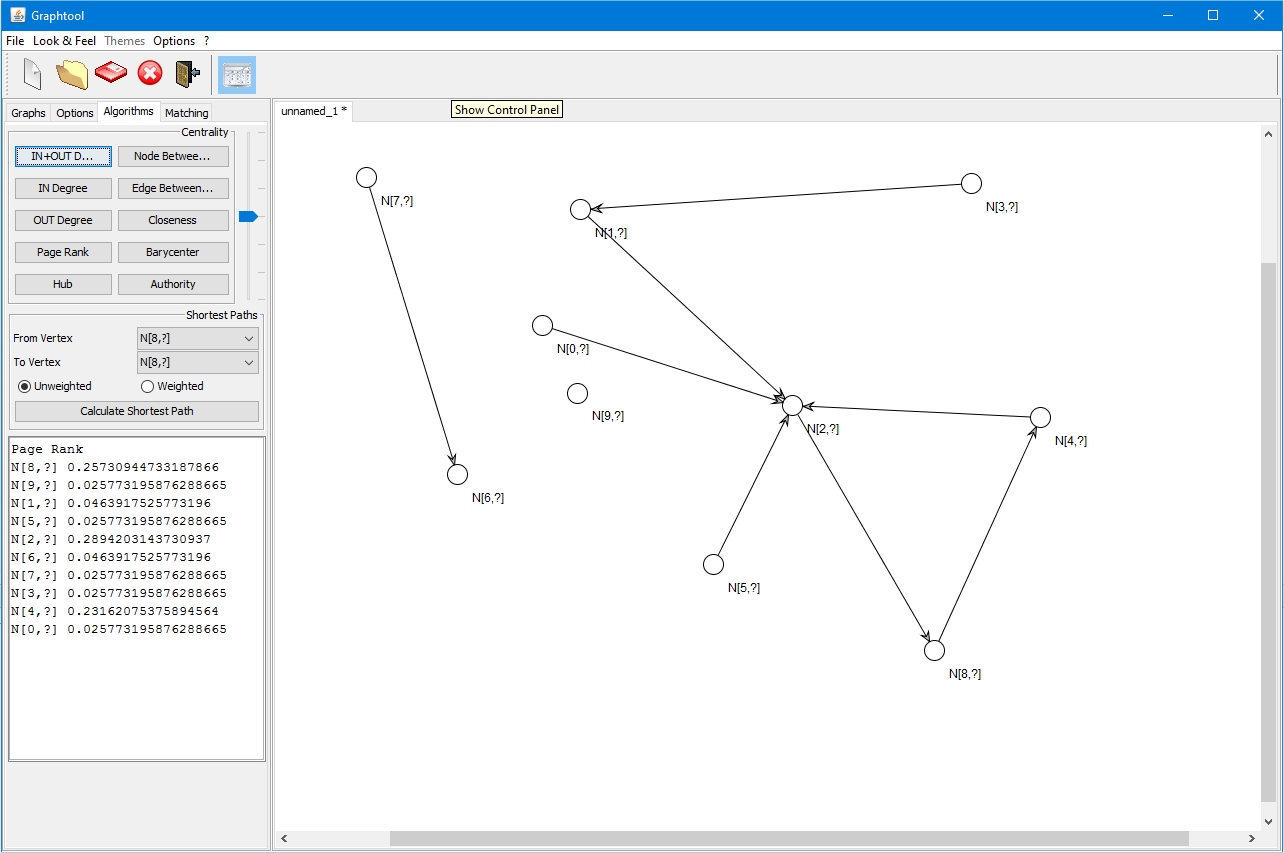
Grapohtool is a softwarre for manipulating graphs and apply several type of algoriothms:
- Bipartite Minimum Weight Matrching;
- Bottleneck Matching;
- IN/OUT degree;
- Betweenness;
- Page Rank;
- Hub & Authority;
- Dijkstra Shortest Paths;
- ...and much more;
Graphtool is supported by StudioPigola on all editions of Windows 10.
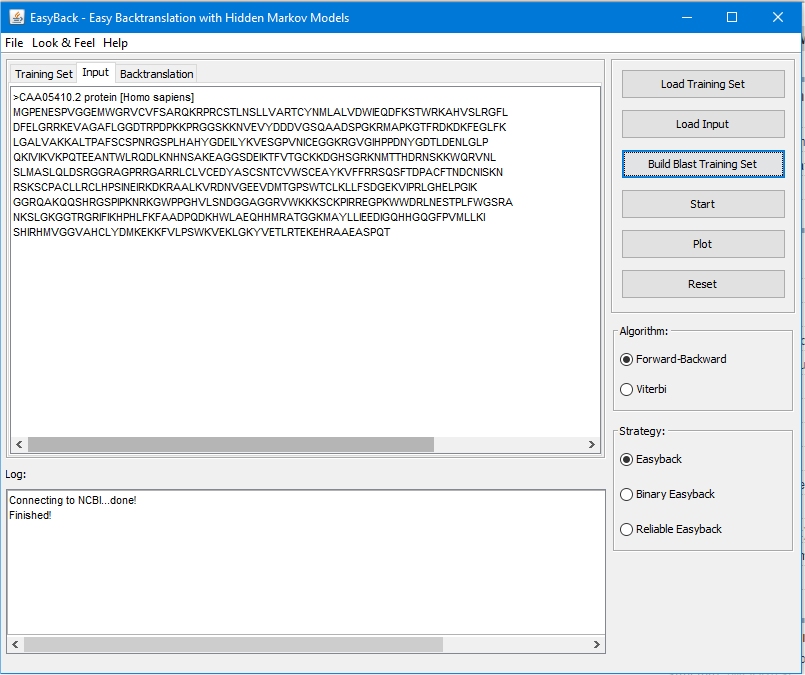
Backtranslation is the process of decoding a sequence of amino acids into the corresponding codons. All synthetic gene design systems include a backtranslation module. The degeneracy of the genetic code makes backtranslation potentially ambiguous since most amino acids are encoded by multiple codons. The common approach to overcome this difficulty is based on imitation of codon usage within the target species. EasyBack is a new parameter-free, fully-automated software for backtranslation using Hidden Markov Models. EasyBack is not based on imitation of codon usage within the target species, but instead uses a sequence-similarity criterion. The prediction quality of a protein backtranslation methis markedly increased by replacing the criterion of most used codon in the same species with a Hidden Markov Model trained with a set of most similar sequences from all species. Moreover, the proposed method allows the quality of prediction to be estimated probabilistically.
This tool has been eveloped with other Autors as a research project at University of Catania.
EasyBack is supported by StudioPigola on all editions of Windows 10.
REFERENCES
- 2007 A. Ferro, R. Giugno, G. Pigola, A. Pulvirenti, C. Di Pietro, M. Purrello, M. Ragusa, “Sequence similarity is more relevant than species specificity in probabilistic backtranslation”, BMC Bioinformatics 8 : 58, 2007. DOI:10.1186/1471-2105-8-58
Synonymous mutations might be involved at the HIV RNA level in HIV-1 drug resistance mechanisms. This tool permits to detect authomatically synonymous mutations.
This tool has been developed in collaboration with University of Rome Tor Vergata, University of Modena and Reggio Emilia, Italy, NMI ‘L Spallanzani and University of Catania.
MutationFinder is supported by StudioPigola on all editions of Windows 10.
REFERENCES
- 2008 V. Svicher, C. Alteri C, A. Laganà, G. Pigola, S. Dimonte, C. Mussini, F. Forbici, A. Ferro, P. Narciso, A. Antinori, F. Ceccherini-Silberstein, C. F. Perno, “Synonymous Mutations are Involved in the Highly Ordered Regulation of NRTI Resistance”, XVII International HIV Drug Resistance Workshop – Melia Sitges, Spain, June 10-14 2008. Antiviral Therapy 13:S3A62 (2008).